
RDF data integration and visual SPARQL query interface
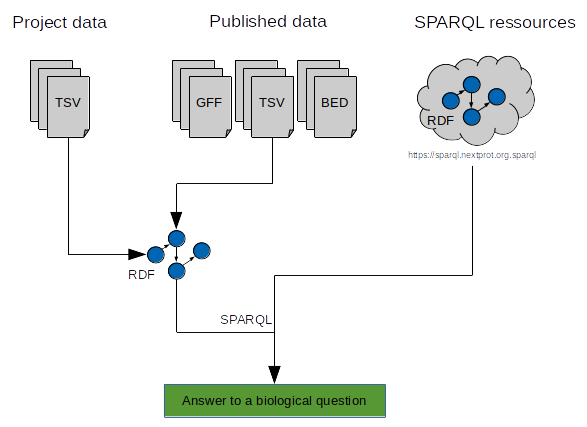
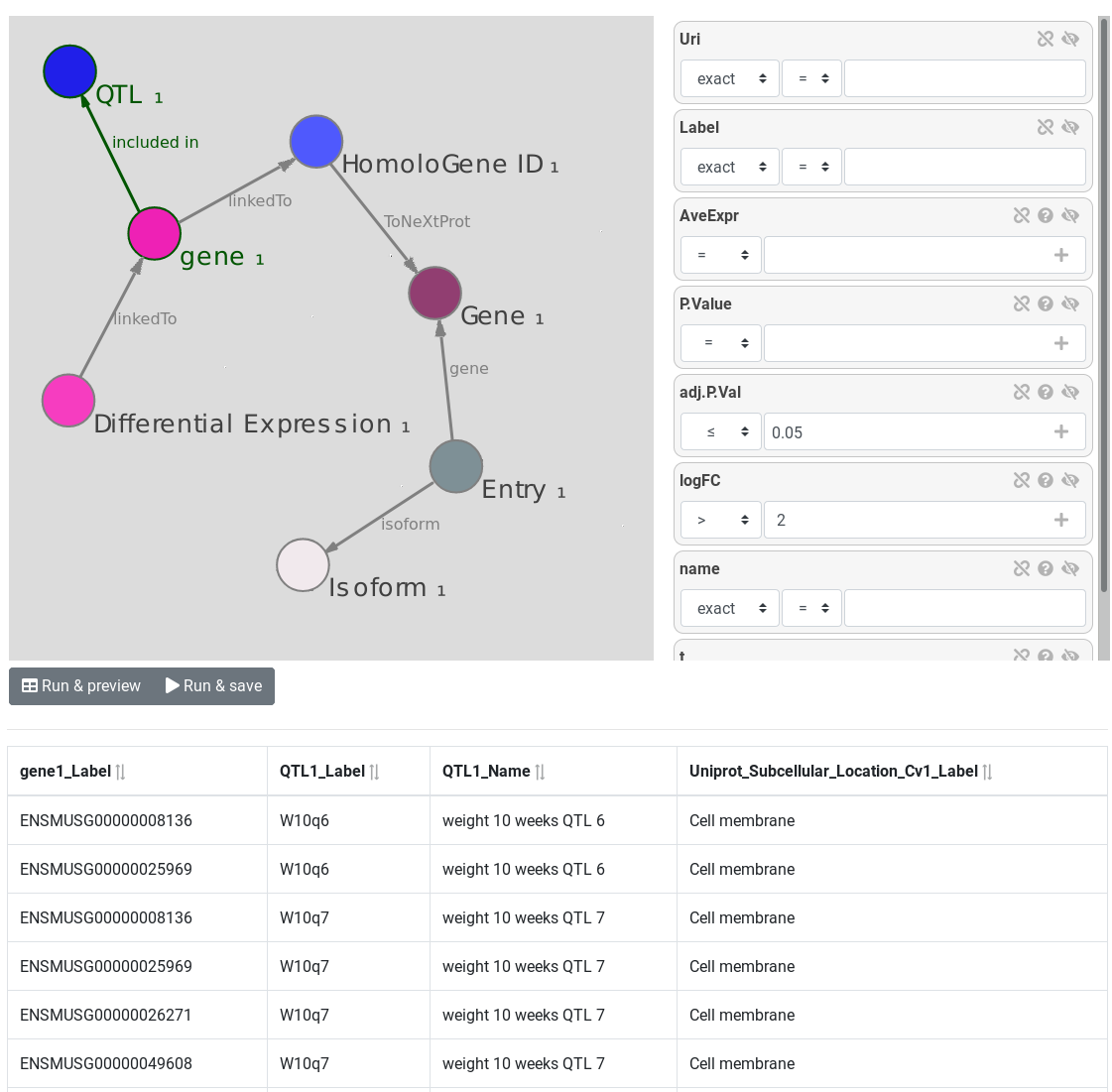
AskOmics aims at bridging the gap between both end user project-specific datasets and with reference databes and knowledge bases from the Linked (Open) Data cloud. It allows heterogeneous bioinformatics data (formatted as tabular files, GFF format or native RDF files) to be loaded into an RDF triplestore and then be transparently and interactively queried through a user-friendly interface.
 |
 |
Interested in using AskOmics?
- The RNA-Seq analysis with AskOmics Interactive Tool Galaxy tutorial is a good entrypoint to learn the basics of AskOmics
- The AskOmics tutorials will teach you how to use AskOmics with your own data
- The deployment documentation allow you to host your own AskOmics instance
- Abstractor can connect your instance with external SPARQL ressources
AskOmics is a free and open source software under the AGPL-3.0 License. All the sources are hosted on the AskOmics GitHub organization.
We acknowledge the GenOuest bioinformatics core facility for providing the computing infrastructure. And the research teams of the authors of this project: Dyliss, Genscale and Igepp.
 |
 |
 |
 |
 |
 |
